Stagerunner
Experimenting with a data science task is usually done through the R console or an IDE by executing portions of a file or collection of files. Sometimes this is slightly generalized into notebooks, interactive sessions that record the history of the code you executed and its outputs, including plots, data, and summaries.
This kind of workflow works well in the old way, when you don't intend on returning to your analysis for any future use and are happy with a static image result.
Developers working on pure software engineering projects usually operate in a different way: they manipulate a single codebase and provide iterability and experimentability through some other approach that ultimately ends up reflecting what's in their codebase.
For example, a front-end web developer might change a JavaScript file in the deep innards of a web app, hit refresh on their browser, and test out the new functionality by clicking their mouse.
To achieve a better data science workflow we will mirror the effectiveness of the typical developer approach. We will rely on a shared codebase composed of modular and testable resources that allows us to go back in time using version control and have new components vetted using code review.
Let's try putting the pieces of the codebase together into a living breathing object, the same way a web browser brings a web application alive. We'll start with a stagerunner but in the future may end up with much more.
The Execution Cycle
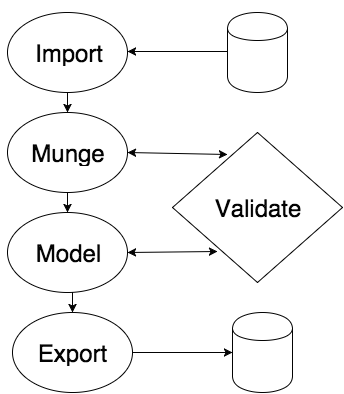
As mentioned earlier, a typical data science cycle begins with: importing data, munging (cleaning) it, running a statistical classifier, and exporting the model for validation and deployment.
A stagerunner is a single R object (specifically, an R6 object, for those familiar with the R6 package which augments R's object-oriented programming capabilities) that represents the full execution cycle of the data science process. In workflows with large data sets, this process is typically distributed across many machines in frameworks like Spark and Hadoop. Since there is no distributed implementation of the R interpreter (yet - shh ;) ) we focus for now on small and medium size data problems. Typically, productionizing several 80% solutions offers more business value than productionizing one 100% solution and anyway downsampling is often good enough.
A stagerunner is simply a tree structure whose terminal nodes are functions that operate on an R environment object. If you are unfamiliar with R environments but understand pointers from languages like C++, simply think of it as a pointer to a list. These functions are executed linearly on the R environment starting with the empty environment and culminating in side effects that export a model, such as storing it to a backend like a cloud storage service or a file. For the mathematically inclined, think of a stagerunner as an element from finite sequences on the monoid of endomorphisms of R environments with the additional meta-data of a hierarchical structure (a very fancy way to say a list of functions that take and return exactly one environment)
Some task scheduling frameworks like Luigi and Airflow take arbitrary directed graphs of tasks and this is certainly the correct generalization but can increase the difficulty of debugging failures in complex topologies of tasks. Staying within a console tends to be much cozier. Eventually Syberia may release this kind of hybrid in-console task scheduling and debugging but for now let's master stagerunners. (If you think hard enough about it, any directed graph can be decomposed into purely serial components.)
A simple stagerunner
Before we jump into the convenient conventions set up by the modeling engine that ships with Syberia by default, let's review the basics of stagerunners.
r <- stagerunner$new(list2env(list(data = iris)), list(
"Double first columns" = function(env) { env$data[[1]] <- 2 * env$data[[1]] },
"Drop categorical columns" = function(env) { env$data <- Filter(Negate(is.factor), env$data) }
))
r$run()
Ok so you got me, I lied a little. Stagerunner functions do not need to return the environment they modify since this is one of the few R objects that is passed by reference and not by value, and hence any modification in another scope (like a function call) modifies the original environment object.
Observe that running the above code yields the desired changes:
str(r$context$data) shows we have a modified
version of iris with the first column doubled
and the only categorical column removed.
env <- list2env(list(data = iris))
r <- stagerunner$new(env, list(
"Double first columns" = function(env) { env$data[[1]] <- 2 * env$data[[1]] },
"Drop categorical columns" = function(env) { env$data <- Filter(Negate(is.factor), env$data) }
))
r$run(1); r$run(1); r$run(1)
Inspecting head(env$data[[1]]) shows that the first column of
iris is octupled as expected. Now try re-executing all but
the last line and type r$run(2). The last column
was dropped, as expected.
This is an example of a stateless stagerunner: it has no memory about which stages ("Double first columns" versus "Drop categorical columns") were executed. Typically this matters as many feature engineering operations are not commutative: dropping correlated features and then imputing may yield different results than doing it the other way around.
More complicated runners

To enforce the order of operation and gain an additional feature we enable stateful stagerunners. This unlocks the ability to replay any part of the data science process like checkpoints in a video game.
r <- stagerunner$new(list2env(list(data = iris)), list(
"Double first columns" = function(env) { env$data[[1]] <- 2 * env$data[[1]] },
"Drop categorical columns" = function(env) { env$data <- Filter(Negate(is.factor), env$data) }
), remember = TRUE)
r$run(2)
Note the remember = TRUE flag at the end. The above
code will fail with "Cannot run this stage yet because some previous stages have not been executed."
This is because we are trying to run step two without first running step one.
Running r$run(1) first will fix the problem.
env <- list2env(list(data = iris))
r <- stagerunner$new(env, list(
"Double first columns" = function(env) { env$data[[1]] <- 2 * env$data[[1]] },
"Drop categorical columns" = function(env) { env$data <- Filter(Negate(is.factor), env$data) }
), remember = TRUE)
r$run(1); r$run(1); r$run(1)
Inspecting head(env$data) shows that the first column
of iris was doubled, not octupled! This is because
with remembrance of state turned on our stagerunner object begins with
a cached copy of the original environment rather than operating on the
latest environment. This is powerful: with stateful stagerunners, we can simulate
going back in time to any step in the process!
The code behind stagerunner makes some space-time tradeoffs. With stateful runners, a full copy is made of the environment on each step. This is prohibitively memory expensive with large datasets. With small datasets, it is manageable and gives us fantastic interactive data analysis and debugging capabilities.
The typical workflow within the Syberia modeling engine is to use an R option or flag to import a small subset (a couple thousand rows) of the desired data set and develop a full-fledged prototype of a feature engineering pipeline and classifier. Any immediate errors or unexpected bugs in feature engineering can be rapidly debugged by effectively using stagerunners.
When the data scientist is satisfied with the end-to-end process, she simply flips the stagerunner to run in a stateless context on the full data set and gets to have her cake and eat it too; no more painful debugging of background batch jobs or digging through logs.
A final goodie: by using objectdiff, we can reduce the space-time tradeoff even further by only copying the elements of the environment that changed during each step. If only one column was doubled in a thousand-column dataframe, the stagerunner and objectdiff combo heuristically analyzes the output of the functions that are executed and rapidly detects only one column was changed, minimizing how much is stored in the "git-like patches" recording the differences in each stage. This retains the replay capability of stagerunners while minimizing memory overhead.
env <- objectdiff::tracked_environment(list2env(list(data = iris)))
r <- stagerunner$new(env, list(
"Double first columns" = function(env) { env$data[[1]] <- 2 * env$data[[1]] },
"Drop categorical columns" = function(env) { env$data <- Filter(Negate(is.factor), env$data) }
), remember = TRUE)
r$run()
Note that class(env) == c("tracked_environment", "environment").
Inspecting objectdiff::commits(env) shows that two commits
have been made and running objectdiff::rollback(env, 1) shows
that ncol(env$data) == 4.
It's possible to nest stagerunners:
r <- stagerunner$new(new.env(), list(
"Import data" = function(env) { env$data <- get(get("DATASET", envir = globalenv())) },
"Munge data" = list(
"Double first columns" = function(env) { env$data[[1]] <- 2 * env$data[[1]] },
"Drop categorical columns" = function(env) { env$data <- Filter(Negate(is.factor), env$data) }
),
"Make a model" = function(env) { env$model <- stats::lm(Sepal.Length ~ ., data = env$data) }
))
DATASET <- "iris"
r$run() # Note that class(r$context$model) is an lm object.
The modeling engine
The default Syberia engine provides a lot of syntactic sugar to make the above process less painful. In particular, we make the notion of stages primitive and parametrizable: a stage is simply one node of the full execution tree of a stagerunner. The top-level stages that come out-of-the-box are import, data, model, and export stage but it is trivial to create more.
The power of this approach lies in finding good parametrizations of the data science process. Syberia is a meta-framework that allows you to build DSLs customized to solving your specific data problem. To use it correctly you may have to create new abstractions (like finding the correct parametrization for your stages) in order to reap its full power.
Web frameworks like Rails discovered that the model-view-controller pattern works well for many web applications, but as data science is a field still in relative infancy, it is your and the Syberia commmunity's duty to explore new paradigms for rapidly iterating on data science solutions.
Enough hype. Let's see an example of an actual data science workflow with this approach.
# Open up the example.sy project from earlier and place this file in
# models/dev/example2.R
list(import = list(R = "iris"))
Under the hood, typing run('example2') creates a stagerunner
that is roughly equivalent to the following code:
import_R <- function(params) {
get(params, envir = globalenv())
}
import <- function(params) {
adapter <- names(params)[1]
params <- params[[1]]
adapter <- get(paste0("import_", adapter))
adapter(params)
}
r <- stagerunner$new(new.env(), list(
"Import data" = function(env) { env$data <- import(list(R = "iris")) }
))
r$run()
That seems like a very complicated way of saying env$data <- iris!
It is. However, we have gained something in the process. Note that our model file
is simply one line of code and clearly reads "import data from an R global variable
called 'iris'." If we replace it with the below code we would read from an Amazon S3
cloud storage key without any additional work.
list(import = list(s3 = "some/s3/key"))
It is possible to create custom import adapters that read from your production database, a data warehouse, a stream, etc.
The modeling engine includes a few very minimal adapters but we will later see how to create your own.
list(
import = list(R = "iris"),
data = list(
"Create a dependent variable" = list(renamer, c("Sepal.Length" = "dep_var")),
"Drop categorical features" = list(drop_variables, is.factor)
)
)
After run("example2") take a look at
head(A) and you will see that we have a
dep_var column and Species has
been dropped. The above shows a parametrization of feature engineering:
instead of writing out the full code for renaming the variable and
describing in R code how to drop the factor columns, we have
used a shorthand for describing mungebits to achieve our
feature engineering pipeline.
list(
import = list(R = "iris"),
data = list(
"Create a dependent variable" = list(renamer, c("Sepal.Length" = "dep_var")),
"Drop categorical features" = list(drop_variables, is.factor)
)
)
It is trivial to add new import and export adapters, parametrize mungebits that do complex feature engineering tasks, and parametrize statistical classifiers using any R package from CRAN or GitHub.
list(
import = list(R = "iris"),
data = list(
"Create a dependent variable" = list(renamer, c("Sepal.Length" = "dep_var")),
"Create a primary key variable" = list(multi_column_transformation(seq_along), "dep_var", "id"),
"Drop categorical features" = list(drop_variables, is.factor)
),
model = list("lm", .id_var = "id"),
export = list(R = "model")
) # Try typing class(model) after run("example2")
The default implementation of model stage requires a primary key in the data set so we artificially create one. This is so we can easily remember train/test splits when making production models on real data sets later. The default model stage encourages storing train IDs on the model object by deisgn. You almost always want to remember the train IDs so you can perform validation correctly in the future and correctly compare refits against past models without predicting on training rows.
With the Syberia modeling engine you enter the realm of developers and are no longer playing in an interactive notebook but recording the fruits of your labor into a breathing codebase with the aid of a very interactive execution environment. In its ideal incarnation, Syberia deprecates the need for notebooks and other data science tools and replaces it with a simple process understood by all software engineers: write clean, modular, and testable code so that anything you want to create is at your feet. If you are smart enough to master the statistics behind data science then you can leave the IDE behind and learn the development tools that make software engineers so capable and productive. You'll also increase your salary.
Re-running stages
The real power from the stagerunner approach lies in replay capability. The standard development cycle for a new model should proceed as follows.
- Determine what data you want to import. Ensure your adapter has an easy toggle for importing a tiny subset of the data (e.g., you should be able to restrict to a few thousand rows by setting a certain global option).
- Build your feature engineering pipeline, replaying each step and
checking the
BandAglobal variable helpers to examine what the data looks like before and after running a munging step. - Train a small model and check that the parameters are sane.
- Run a full model by flipping the aforementioned toggle and make sure to turn off the remember flag on the stagerunner.
This is a local development workflow. The last step should be executed on a remote build scheduling system that deploys instances capable of handling your large data sets (like a large Amazon AWS instance or a multi-instance operating system like Plan 9--just kidding, but that would be cool). Alternatively, you can SSH into a large instance and run models there directly but it will be cumbersome to try experiments that simultaneously train thousands of models.
For example, try the following commands.
run(, 1)
run(, "2/1")
run(, "data/2")
run(, "da/1", "da/2")
run(, "data")
run(, "model")
run(, 3)
Carefully inspect the structure of B and
A before and after each command.
Note that these helpers are really active bindings and
boil down to r$context$data on the
underlying stagerunner that is being constructed on-the-fly
before and after calling r$run("2/1"), etc.
The extra comma at the beginning indicates we wish to continue to
run the "example2" model. The difference is that we are calling the
run global helper which takes an additional first parameter rather than the
run method on the underlying stagerunner. Typing run("example1", to = 2)
indicates we wish to switch focus to the "example1" model and execute its
process up to the data munging stage.
Note the fuzzy matching in the above examples. Under the hood,
"da/1" means "look for a stage that matches the regular expression
.*d.*a.* in its top-level and grab the first node." What
happens when you try run(, "d/1")?
An important note is that fuzzy matching can be ambiguous and
sometimes execute the wrong model. Fuzzy match results are ordered
in descending order by file modification time, so if you ever find
run is being annoying and not executing
the correct model (e.g., if you have "example" and "example_foo"
and run("example") is matching to
the incorrect one) simply make a tiny change like adding a space
and save the model file: run will
pick up the change and execute the model whose file has been
most recently modified.
The modeling cycle
You now know enough about stagerunners to start being productive. The next step is to learn more about how the various stages are parametrized and learn how to extend them and write your own. For example, if you need to generate a report after the model is trained but before exporting the object, you could write a "report" stage that creates an appropriate R markdown file. Or maybe you want to deploy a web service that has a dashboard after you have exported the model: simply write a "dashboard" stage.
To summarize, we have learned that:
- Model files are simply syntactic shortcuts for building stagerunner objects. Typically, you never have to explicitly deal with these objects.
- Stagerunner objects are syntactic shortcuts for executing parts of a list of R functions that take and modify an environment called the stagerunner context. In other words, stagerunners express in code the idea of highlighting a section of an R file and hitting the execute button.
- The power of using stagerunners derives in our departure from messy, linear scripts. By separating components into clearer functions, we can let duo's like stagerunner and objectdiff memorize the changes that we make to our data so we can replay our steps and debug what went wrong.
- Stages are functions that take parameters and produce a list of stagerunner-compatible functions (re-read that a few times!). By defining a good set of parameters for the data science process and splitting it up into stages like import, data, model, export, and more, we gain the power to succintly describe how to take a messy, production data set and convert it to a ready-to-deploy model. This succint description is created by the data scientist during interactive data analysis but is complete and admissible into a version-controlled codebase so we never have artifacts or scripts lying around.
- The modeling engine provides lots of shortcuts and shorthand for rapidly developing models and feature engineering pipelines while retaining the flexibility of debugging and testing.
- The best part is: the end result is a clean file in a beautiful codebase without the typical mess left by an interactive notebook or R script! The whole thing forces you to be a good developer by design by reducing the friction in developing data science products like a software engineer.
One last time, enough hype! Let's dive in to a few more features of the modeling engine.
