Syberia Modeling 101
Ensure you understand the basic usage of the modeling engine prior to proceeding.
Developing classifiers using the Syberia modeling engine follows one primary tenet: development equals production. When a web developer experiments with a new website layout or feature, they are targeting a production system. After they are satisfied with their work, they push a button and deploy the code base so it is live and others can interact with it.
Feature engineering and statistical modeling (as we will see, inseparable components of machine learning) should belong to the same class of work. When an architect designs a skyscraper their work has to be translated to reality through a construction team by replaying the design using a physical medium. This is the current industry standard for machine learning: prototype the model in one language, typically a Python or R "notebook," and then write it in a "solid production-ready" language so it can survive the harsh winds of the real world.
This is wrong. Prior to the invention of HTML, construction of UIs was a laborious design process that proceeded from the lab to the factory. HTML, and later JavaScript, introduced a syntax, a grammar, for rapidly experimenting with UIs and quickly pushing them to the real world.
The tenet of using different languages for development and production is fundamentally flawed. Good syntax will always win because it records the critical mathematical abstractions relevant to the domain and retroactive optimization becomes possible. Recoding your hardly fought experimental fruit from R to Scala or from Python to C is a waste. Before HTML, inter-network UIs were written in C and other lower languages. The world wide web only exploded because it gave the power of rapid, iterative UI development to all through a simple and straightforward syntax.
Teaching all the world's statisticians and applied mathematicians C, Java, or Scala to "productionize" their work is not a viable long-term strategy. The only correct approach is to take the syntax, the mathematical notation, most comfortable to these experts and then wielding a new world from it: one that is alive.
Whether any aspect of the current Syberia framework or its de facto modeling engine survives is irrelevant, in the same way that VBScript ended up irrelevant, but it is important to take that first step. Do not be afraid to see real-time production-ready hybrid stream-and-batch data processing and statistical prediction as an experimental process that culminates in a finished product the same way experimenting with a web app culminates in a deployable real world tool without additional code or re-implementation work.
We have bothered with a philosophical prelude because we believe the statistician and scientist should come first: there should be no "data engineer" trained in computer science and difficult-to-master functional languages and distributed systems to rewrite the work from scratch to make it "ready," at least to the same extent there is no "web engineer" that takes a UI/UX expert's JavaScript work and translates it to a C++ application every single time. All this should be the corollary of a well-designed syntax, and the data engineering task should be a one-time fully general abstract undertaking that implements that syntax in the medium of streaming distributed systems. We are trying to find that syntax, and R's homoiconicity and interactive nature forms the perfect testbed for finding that grail.
Naturally, some organizations and performance-intensive applications will continue to require full rewrites of the experimental designs, but there is no reason why a well-designed syntax should not be capable of auto-transpiling to the performance-optimized description, in the same way that assembly languages were subsumed by higher abstract tools.
With all that said, let's jump into what a primordial first iteration of a complete modeling grammar might look like. If you like this approach to statistical modeling, maybe you can help us design its future.
The Basic Modeling Cycle
The first analogy that came to my mind is of immersing the nut in some softening liquid, and why not simply water? From time to time you rub so the liquid penetrates better, and otherwise you let time pass. The shell becomes more flexible through weeks and months—when the time is ripe, hand pressure is enough, the shell opens like a perfectly ripened avocado!
— Alexander Grothendieck
Finding well-designed syntax is hard. Historically, many achievements in math and physics were a direct result thereof. Nevertheless, it is worth doing because a successful execution allows you to think at a higher level of abstraction. We start with the simplest possible grammar so that we may extend and build atop it to achieve more complexity. We will do so without getting distracted by the latest and shiniest database tool or complex ensemble topology, but instead with the meticulous patience of a mathematician, until the formerly daunting seems trivial.
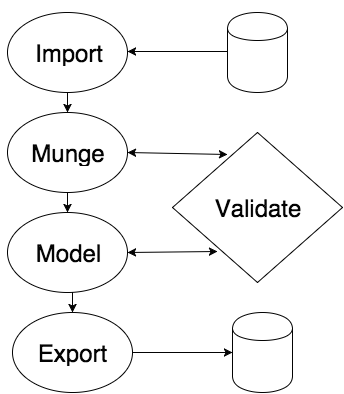
For now, we stick with the standard supervised learning problem on a binary or continuous predictor. Any instance of this machine learning problem requires us to get data to perform analysis on. On the other hand, typically "export" is not embodied as a single button but as a process: re-coding the results of the experimental sandbox analysis into a production codebase. The Syberia approach is to turn this around and demand that all metadata relevant to reproducing the modeling process in a real-time setting be attached to a single serializable object. Fortunately, R makes this easy.
The first grammatic abstraction is a fairly commonplace one: that of
I/O adapters. We need a way to read in data, typically from a file
or a database, and a place to store our results, typically a file or
cloud storage. We will see how defining short snippets of code in the
lib/adapters directory in a Syberia modeling
project allows us to import arbitrary data and export models arbitrarily.
What happens in the middle? Once we have the data, we typically have to clean it. This is because the real world is messy and we rarely get exactly what we expect: we may have missing values, mislabeled points, insufficiently massaged features, and a whole host of other problems. Determining which variables to keep after arduous experimentation can be first-order approximated using sure independence screening. The process of data preparation in the Syberia modeling approach is presently performed using mungebits, atomic production-ready feature engineering templates that allow us to quickly parametrize the end-to-end cleaning process without having to write what usually boils down to fairly repetitive code.
The other, perhaps primary, purpose of mungebits is to encode any metadata we may need to replay our feature engineering steps in production. For example, if we imputed certain columns with the mean, we will have to pre-record the means of each imputed column, as this information will not be available in production when a new record streams in.
After the data is "clean," in the sense that it forms a nearly complete primarily numerical matrix without incorrectly labeled signals, we can try a host of machine learning techniques on it: generalized linear models, support vector machines, neural networks, clustering, random forests, and so on.
Typically, recording the model is done in a format like PMML, but this is too restrictive. It forces us into a very limited set of feature engineering steps and statistical parameters. Instead, we leverage the power of R's LISP-like computation and store any code and data required to reproduce the predictions within a single R object called a tundra container (named tundra because it freezes the model for production use :).
To determine whether we are satisfied with the model, we have to apply some sort of validation. Typically, this may be measuring the AUC of the predictions against a target threshold. Note that both feature engineering and choice of statistical learning parameters may heavily influence performance.
Together with importing data and exporting results, the iterative process of experimenting with feature engineering and statistical classifier parameters forms the heart and intersection of most approaches to modeling problems, so we will focus on solving this grammatical exercise until it becomes a trivial feat before proceeding with higher grammars.
Throughout, we aim to make the description of the modeling process as concise as possible while retaining the tip of the iceberg composing the critical information. If a model file ever grows to longer than 100 lines of code, it is a good sign you should split it up and refactor. Let's revisit our earlier trivial example.
list(
import = list(R = "iris"),
data = list(
"Create dep var" = list(renamer, c("Sepal.Length" = "dep_var")),
"Create ID var" = list(multi_column_transformation(seq_along), "dep_var", "id")
),
model = list("lm", .id_var = "id"),
export = list(R = "model")
)
The last expression of every model file is a list.
A list itself is a static object, but the assembly
of a list into a living, breathing avatar of the modeling process occurs
internally when typing run("example").
The Four Stages
Let's break down the four stages.
Importing data is done through the import stage. This stage looks at
lib/adaptersto find the necessary adapter. The modeling engine comes with several built-in adapters, including the R, file, and S3 adapters for importing from and exporting to R's global environment, files, and Amazon's S3 cloud storage, respectively. Reading the name of the list element"R"fromlist(R = "iris")allows the import stage to pluck outlib/adapters/R.You can define a new way to import data by placing a file with
readand/orwritelocal variables holding functions inlib/adapters/your_new_adapter.Rand then replacing the first stage withimport = list(your_new_adapter = ...)where the right-hand side is the parametrization that will be passed to thereadfunction. Later, we will examine further how to do this (and test that it works as expected).Munging the data is achieved through the data stage. Each stage accepts its own parametrization and convention. While the import stage is relatively simple, describing merely the external location of the data, the data stage can be as complicated as you need it to be.
The vanilla data stage is parametrized by a named list, with each list element name representing a very brief summary of the feature engineering or cleaning step, and the list element value representing a grammar of data preparation we will explain down the road.
The nice thing about this grammar is that it allows us to write almost any operation on the data set in one line of code and separate the abstract parametrized code for performing the operation into a separate place,
lib/mungebits, where all of its inputs and outputs can be thoroughly tested and ensured of correctness.Running the model! This step is frequently and incorrectly viewed as the most important component of the modeling process: the secret sauce is in finding the correct parameters to a magical mathematical method. This is wrong. Once you start writing some Syberia modeling files, you will see that the meat often lies primarily in the data preparation, not in the statistical method. It is a pity that most books on machine learning do not make this acknowledgement.
The parametrization for the model stage is simple: a list whose first element describes the name of the classifier we will use, which convention dictates it fetches from
lib/classifiers/lm.R. Any file in that directory will be a short R file containing two local variables,trainandpredict, where the former records the metadata required to play the predictions on any subsample of the original dataset (in particular, single rows of streaming data). Typically, this is what we refer to when we say we are estimating the "parameters" of a statistical learning function.The rest of the list elements will be fed directly to the
trainfunction after the data has been cleaned. The Syberia modeling engine provides the shortcutMto represent the model object so that we can preview some predictions usingM$predict(validation_data).Hand-in-hand with some function
validate(M), we can downsample the import stage prior to experimentation and use the re-running functionality to playrun(, 3)(i.e., re-run the model stage) to make a succession of plots or estimates of the efficacy of various parameters, before settling on a choice for the final dataset. This is a typical approach to experimenting with data sets that fit into memory, but there is a way to make it work on arbitrarily large data sets as well.Exporting the model. Borrowing from the adapters introduced in the import stage, this stage follows a similar parametrization: the name of each list element should correspond to an entry in
lib/adapterswith awritelocal variable determining how to save the model object based off the parametrizations provided on the right-hand side.In our trivial example, we exported to the R global environment so we can inspect our model. If we are happy, we could add
file = "~/tmp/model.RDS"(or even better, use an S3 key) indicating we are using thefileadapter to export the model object for later use. Typingrun(, 4)will re-run the export stage with this new export adapter, recording our model permanently.
The example we described is a trivial one. What if we wanted to do something more complicated? Imagine we would like to import data from a database, run three complex feature engineering steps (say principal component analysis, some time series aggregation, and sure independence screening), followed by running a model with a new package we found on CRAN, and then exporting the serialized object to a Mongo database.
How much work would we have to do?
# models/dev/fancy_model.R
list(
import = list(database = list(table = "my_data")),
data = list(
"Aggregate time series" = list(aggregate_ts, vars = c("timevar1", "timevar2")),
"Sure indep screening" = list(sis, pval_threshold = 0.05),
"Principal components" = list(pca, center = TRUE, scale = TRUE)
),
model = list("fancy_model", param1 = 0.7, param2 = "foo")
export = list(mongo = "models/experimental/fancy_model/1")
)
- We would have to write a
database.Randmongo.Rfile with areadandwritemethod, respectively, placing them inlib/adapters. (And write tests! More on this later.) - We would need
sis.R,aggregate_ts.R, andpca.Rinlib/mungebits. We will examine later what goes into writing mungebits, but it isn't much. - We would need to wrap the CRAN package in a
lib/classifiers/fancy_model.Rfile.
In total, we had to touch six files, excluding the model file, with some huge benefits: (1) as we will see later, each of our files requires tests for the project to pass continuous integration, ensuring we are confident the results are correct, and (2) we have made the fruits of our work accessible to anyone else working on the project!
This latter point is crucial:
if we had written a linear, serial script, someone would later have to go
dig through our model, potentially sprawled across several files,
to figure out what to copy and where to inject new parameters to
appropriate to their particular data set. If six months later we have written
100 models that used pca and discovered
a subtle bug, we could fix all models simultaneously instead of wading
through old files or notebooks that will inevitably be forgotten.
Forget notebooks. By thinking like developers even when experimenting, we can grow stronger. We can have the best of both worlds. By writing the modeling process in this way, the Syberia modeling engine is forcing the work we write to be modular, testable, and reproducible, preferably in a versioned codebase that records all of history.
This is what gave great power to frameworks like Ruby on Rails, tools that were not hailed theoretical results yet empowered formerly siloed developers to harness the benefits of convention over configuration and reach developer productivity and project iteration speeds that were formerly inaccessible.
The dream of the modeling engine is to reach and surpass this level of productivity as applied to the domain of, well, whatever it is that R does! Machine learning is only the beginning.
